
Charles Chapple
Head of Bioinformatics
On 9th June 2023 we will be releasing the Q2 API changes for the VarSome API (stable-api.varsome.com).
The Stable API will update version 11.6.1 to version 11.7.2 with data frozen as of the 12th May 2023. This version has been successfully used in production for over a month.
The most recent version, used by VarSome itself, is always available at api.varsome.com.
Functional Changes
The major changes in this version were introduced in version 11.7
- Overall improvements:
- Various in-silico predictors improvements
- Extension of HGVS notation supported by VarSome to include single amino acid deletions and duplications within the same exon
- Information from the Deafness Variation Database from the University of Iowa is now available in VarSome
- Germline classification:
- Integration of LOVD evidence as a new source available to Premium users
- Modifications in the way we filter gnomAD improvements
- Further inclusion of ClinGen recommendations for mitochondrial variants
- New in-silico pathogenicity predictor for splice sites, MaxEntScan
- Somatic classification:
- The somatic classifier no longer uses Cosmic
- The somatic classifier no longer uses Cosmic
Please see the version 11.7 Release note for more detailed information.
Database Updates
The following databases have been updated:
|
Source |
Data Type |
Current (Stable, Q1 '23) |
Next Release (Staging, Q2 '23) |
Comment |
|
AACT Clinical Trials |
gene |
19-Jan-2023 |
21-Feb-2023 |
|
|
ACMG classifier |
variant |
11.6.1 |
11.7.2 |
|
|
AMP classifier |
variant |
11.6.1 |
11.7.2 |
|
|
ClinVar |
variant |
5-Feb-2023 |
3-Apr-2023 |
|
|
ClinVar |
region / CNV |
05-Feb-2023 |
03-Apr-2023 |
|
|
DGI |
gene |
19-Jan-2023 |
21-Feb-2023 |
|
|
Genomic England PanelAPP |
gene |
20-Jan-2023 |
22-Feb-2023 |
|
|
GHR |
gene |
19-Jan-2023 |
21-Feb-2023 |
|
|
gnomAD |
variant |
3.1.1 |
3.1.2 |
|
|
GWAS |
variant |
20-Jan-2023 |
21-Feb-2023 |
|
|
Jackson Laboratory CKB |
gene |
24-Jan-2023 |
22-Feb-2023 |
|
|
Jackson Laboratory CKB |
variant |
24-Jan-2023 |
22-Feb-2023 |
|
|
LUMC LOVD |
variant |
1-Feb-2023 |
04-Apr-2023 |
|
|
NIH ClinGen |
variant |
19-Jan-2023 |
21-Feb-2023 |
|
|
OMIM |
gene |
24-Jan-2023 |
11-Apr-2023 |
|
|
OMIM |
variant |
24-Jan-2023 |
11-Apr-2023 |
|
|
PharmGKB |
gene |
20-Oct-2022 |
5-Mar-2023 |
|
|
PharmGKB |
variant |
19-Sep-2022 |
5-Mar-2023 |
|
|
Sanger Cosmic |
variant |
v95 |
v97 |
API customers with a valid Cosmic license |
|
Sanger DECIPHER |
region / CNV |
19-Jan-2023 |
21-Feb-2023 |
|
|
Saphetor Known Pathogenic |
gene |
06-Feb-2023 |
02-May-2023 |
All clinical classifications from all sources. |
|
Saphetor Known Pathogenic |
variant |
06-Feb-2023 |
02-May-2023 |
All clinical classifications from all sources. |
|
Pubmed |
variant |
01-Feb-2023 |
03-May-2023 |
JSON Schema Changes
Reminder: all the data structures returned by the VarSome API are documented here.
Variant Annotation Schema Changes
Body modifications
The following request:
POST /lookup/batch/{ref_genome} accepts now an additional parameter, variant_specs
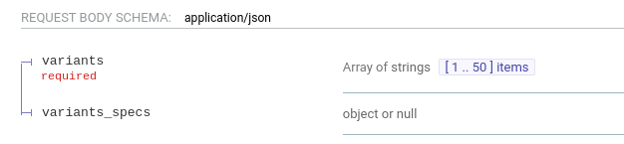
The new parameter is optional and should be ignored by VarSome API users, as it is intended for specific Saphetor installations
Response modifications
The response changes in this section concern both GET /lookup/{spec_query}/{ref_genome} and POST /lookup/batch/{ref_genome} requests.
Attribute removals
Attribute in_silico_thresholds has been removed from the responses of both requests.
Region Database schema changes
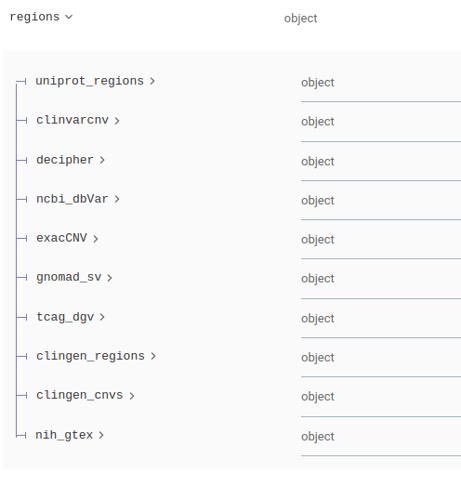
Variant annotation responses under the regions key follow now a specific schema per source database, as is described below.
Region database json keys have also been changed for uniformity:
|
Old name |
Renamed to |
|
NCBI dbVar |
ncbi_dbVar |
|
TCAG DGV |
tcag_dgv |
|
Broad ExacCNV |
exacCNV |
|
NIH GTEx |
nih_gtex |
|
Sanger DECIPHER |
decipher |
|
NCBI ClinVar CNVs |
clinvarcnv |
|
UNIPROT UniProt Regions |
uniprot_regions |
|
Broad gnomAD structural variants |
gnomad_sv |
The regions part of json responses now is standardized as follows in single variant and batch requests:
The detailed response schema per source can be found in the online documentation.
New Databases
The Deafness Variation Database and MaxEntScan database are now present in annotation responses for single variant and batch requests
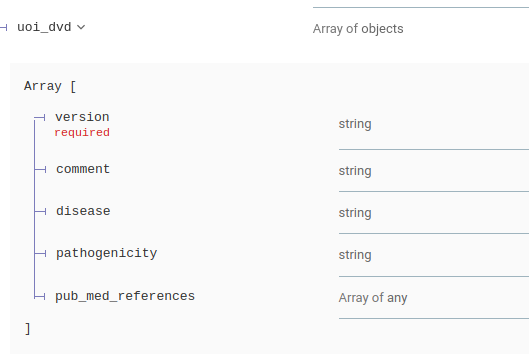
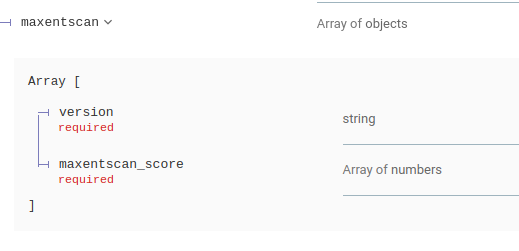
Other Attribute modifications
The pharmgkb attribute in the responses has been modified to include additional information:
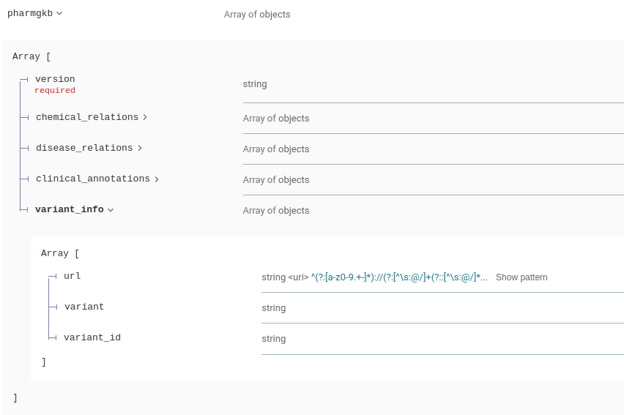
Gene Annotation Schema Changes
Response modifications
The response changes in this section concern both GET /gene/{gene_symbol}/{ref_genome} and POST /genes/batch/{ref_genome} requests.
Attribute modifications
The pharmgkb attribute in the responses has been modified to include additional information:
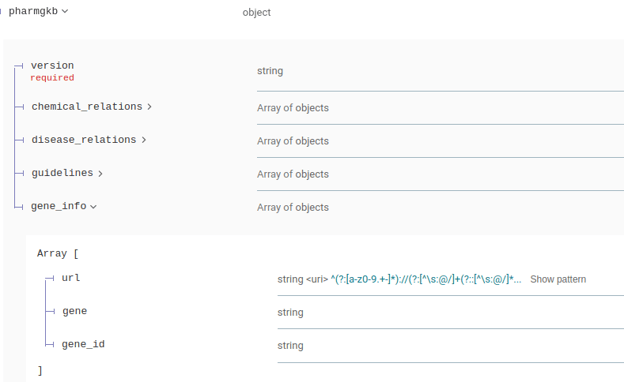
CNV Annotation Schema Changes
Response modifications
The response changes in this section concern GET /cnv/{spec_query}/{ref_genome}
Region Database schema changes
CNV annotation responses under the regions key follow now a specific schema per source database, as described in the Variant Annotation Schema Changes section of this document.
Further Information and Support
An overview of the VarSome API is available here with more detailed information here.
Please contact us at the usual address support@varsome.com should you require any additional information or run into any major issues. As ever we look forward to your feedback and suggestions to improve our platform.

Submit a Comment