In line with the recent releases to VarSome, we are proud to present VarSome Clinical 9.1.
The 9.1 release contains the following major updates, among others:
- The variant calling pipeline has been updated and refined for cancer samples.
- We have additional functionality requested by users, including:
- Shared Filters
- Improved the way gene lists are created from phenotypes
Somatic Variant Calling Pipeline
With this release, we are moving to a different variant caller for somatic samples. We were previously using Sentieon’s TNscope and will now be using Sentieon’s tnhaplotyper2 algorithm. Tnhaplotyper2 is designed to behave like GATK’s mutect2, and our testing has shown that it can sometimes find variants missed by TNscope and also allows us to do better variant filtering.
Tnhaplotyper2, like mutect2, has associated filtering tools which are applied to the variants they find. These filters can then be used to decide whether a variant should be marked as PASS or FAIL. If a variant fails any of the filters present in the “FAIL” column of the table below, it will be marked as FAIL. Failing to pass a filter in the “PASS” column will not cause the variant to be marked as FAIL.
|
PASS |
FAIL |
|
clustered_events |
map_qual |
|
duplicate |
base_qual |
|
fragment |
contamination |
|
multiallelic |
weak_evidence |
|
n_ratio |
low_allele_frac |
|
orientation |
normal_artifact |
|
position slippage |
map_qual panel_of_normals |
|
haplotype |
strand_bias |
|
germline |
|
|
strict_strand |
New software versions
We have updated all external tools used in VarSome Clinical and now have the following:
- GenomeAnalysisTK 4.1.9
- Samtools 1.11
- Bcftools 1.11
- Picard-tools 2.24
- Fastqc 0.11.9
- Sentieon suite 202010
- Delly SV caller 0.8.7
- cutadapt 3.1
- ExomeDepth (R package) 1.1.15
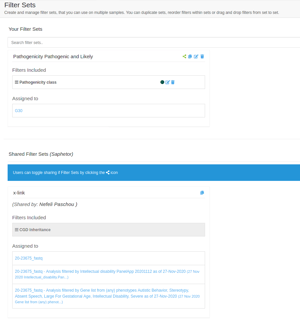
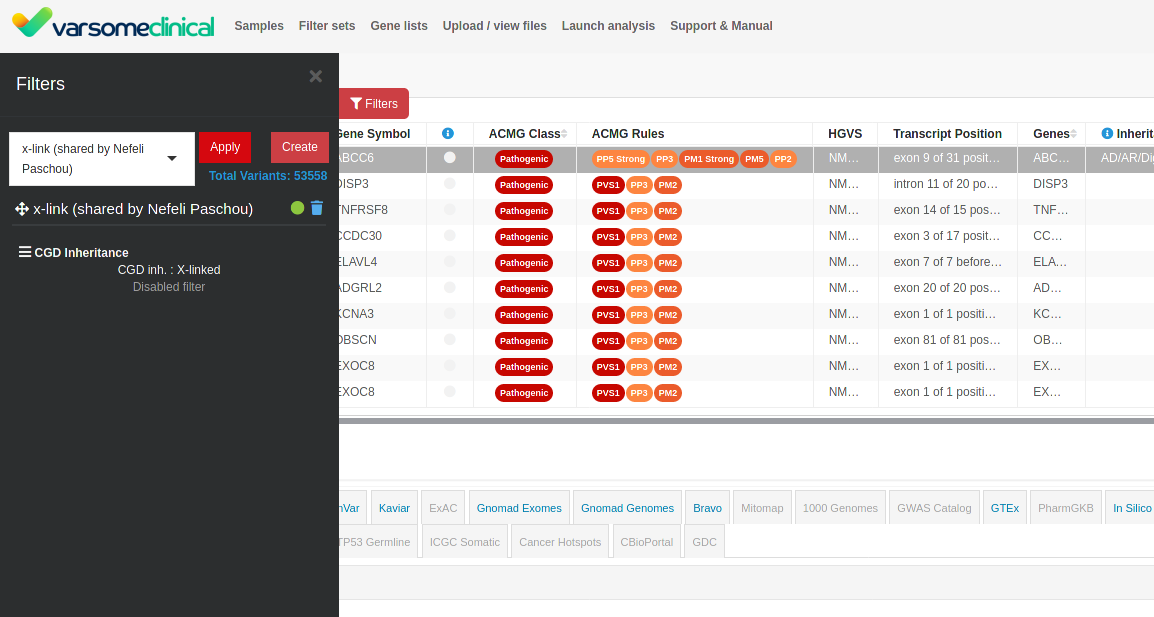
Shared Filters
Users of the same group can now share filters among them. To share a filter with other members of your group, click the Filters link at the navigation bar and then click the share icon on any of the filters you wish to share.
Filters that have been shared among members of the same group appear both on the Filters screen and when filtering a specific analysis. Although a shared filter can be used by anyone in the group, it can only be edited by the user that initially shared the filter. Other group members can still duplicate the filter and edit it.
Gene lists from phenotypes
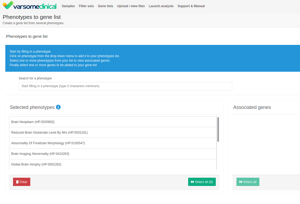
The methodology to generate a gene list from phenotype(s) has been changed. Previously, when making a gene list from phenotypes, we would include only those genes that are directly annotated with that phenotype. We have now extended this and instead first collect all diseases linked to the phenotype and then all genes linked to those diseases, as well as any genes directly linked to the phenotype. We already worked this way when adding phenotypes to analyses, so this change ensures we are consistent and also makes sure we don’t miss any genes when creating gene lists.When generating a gene list from phenotypes or diseases you now have the option to select (by clicking) multiple items from the matched phenotypes. You can also now remove individual phenotypes / diseases by right clicking on the item you want to remove.
Usability changes
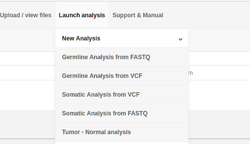
- Launching a germline or somatic analysis has been separated as different options on the user interface.
- Generating a VCF file for sub analyses. You should be able to generate a vcf file with the variants of a sub analysis (gene list, algorithmic filter etc.).
- Reusing component samples of an existing multi sample analysis. You should now be able to reuse component samples of a previous multi sample analysis to initiate a new multi sample or cnv analysis
Cancer Databases
A number of new cancer databases are now available in Clinical, similarly to the free version of VarSome, these include:
- Cancer Gene Census from Sanger
- cBioPortal
- Cancer HotSpots
- Pharmacogenomic Biomarkers in Drug Labeling from FDA
The way the data is presented has been improved for existing databases like Cosmic, ICGC, GDC, IARC TP53. Instead of long lists of samples, we display the key information as charts, showing survival rates, ages of patients, and histograms of tumor locations & cancer types.
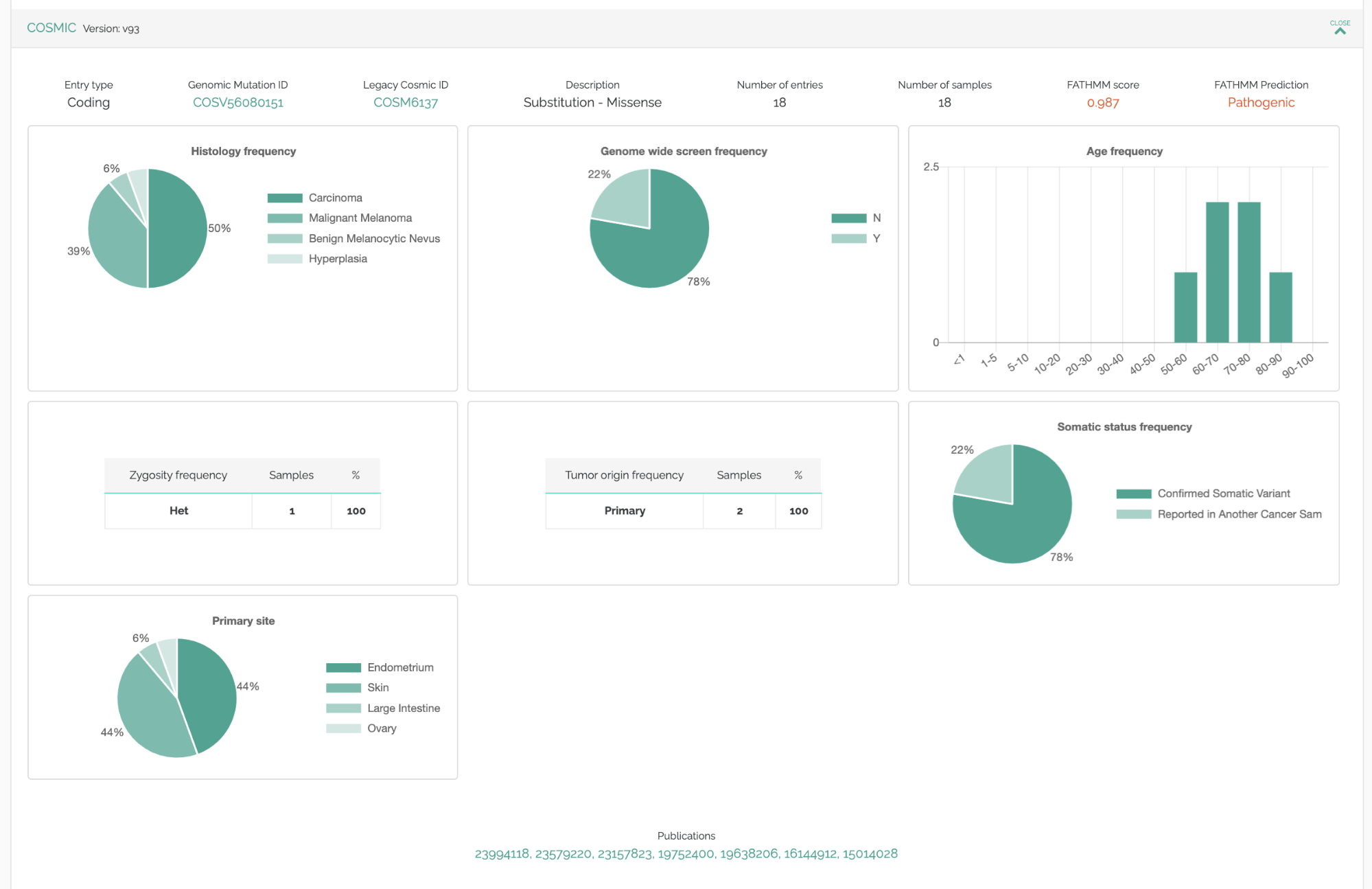
Publication “Tagging”
We have recently introduced an experimental feature which aims to extract important information from the publications available in VarSome: key concepts such as genes, diseases, chemical compounds etc. are automatically highlighted in the text.
ACMG Explanations for failed rules
Full explanations will now be stored for non-trivially benign variants (ie: all non-BA1 variants). This means you can now see the explanations for rules that failed to fire, by simply pressing the blue question mark on the rule that didn’t trigger:

This will help explain sometimes why results may be different from the live free VarSome which doesn’t have certain licensed databases, or may be running a more recent version of the classifier.
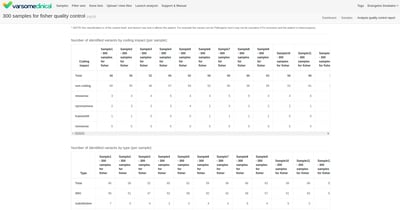
Improved QC Report
QC report has been significantly improved to show the most important information first, and present the data more legibly with better support for large multi-samples.



Submit a Comment