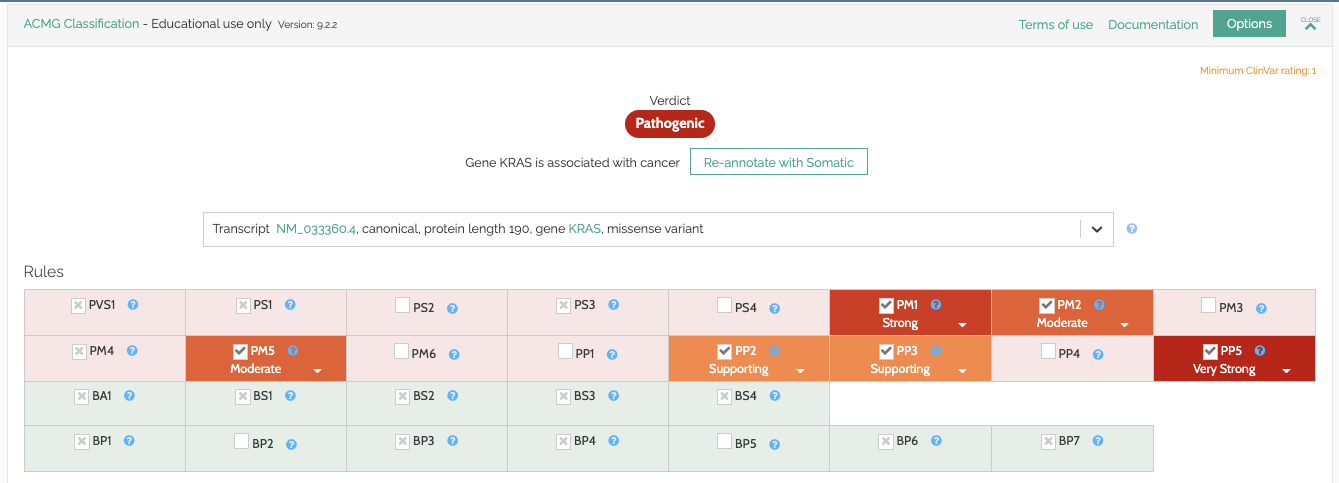
Based on helpful feedback from our users, we have made the following changes to our ACMG classifier in version 9.2.2. In practice, few variant classifications are affected but these changes make our classifications easier to verify and result in slightly fewer false positives.
PVS1: Now uses the observed/expected ratio reported by GnomAD when determining whether a gene has loss of function as a mechanism for disease instead of the Z-Score.
PP2/BP1: Now uses the missense Z-score reported by GnomAD instead of the current implementation that used the ratio of pathogenic/benign missense variants over all non-VUS missense variants.
BP4/PP3: Removed REVEL from in-silico predictions as it is not an independent classification but rather combines the results from other in-silico predictors.
PS1: Fixed an issue that caused some variants to incorrectly double report the same UniProt evidence.
Further Information and Support
Not already a VarSome Premium or VarSome Clinical user? Get in touch and ask for a free trial.
As ever we hope you find these changes & improvements helpful, we’d love to hear any suggestions you may have, support is available as usual from support@varsome.com
- The VarSome Team


Submit a Comment